Abstract
Cooperative forms of animal breeding are still dominating in Southern Germany, Austria and Switzerland. The breeding organizations are supported by public research facilities which allow Bavarian farmers to compete in genetic progress and to apply state of the art methods although the majority of herd book and AI-organizations are way too small to run meaningful research units. The competence centre in Grub comprises a research station, an AI-organization, a veterinary service and research unit, a laboratory for routine genotyping and the head offices of several breeding organizations. The Institute of Animal Breeding runs genetic and genomic evaluations for cattle, pigs, sheep and horses in Bavaria and beyond and has developed cooperation with many other countries in the alpine region. These collaborations are continuing to grow, but have recently been challenged by new developments in international cattle breeding that might also affect the collaboration in genetic evaluation.
Keywords
Animal breeding research, cattle breeding, pig breeding, genomic selection, genetic evaluation, polled cattleCo-Author
Malena Erbe
Introduction
Animal breeding is an important global contribution to meet the growing demand for animal protein. Across the world, different forms of organization have developed, ranging from community based breeding systems in developing countries and cooperative breeding programs across several countries, to global enterprize breeding programs that supply different varieties of their breeding stock to customers in many countries. Which type of breeding program dominates in a certain region depends on the state of development as well as on species, tradition, competition and public support.
The state of Bavaria in Germany has pursued a policy known as „the Bavarian Way“ ever since 1969 that regards rural politics as closely related to social policy and tried to slow down the growth of farming structures while maintaining competitive income and avoiding fallow areas in unfavorable regions [1]. One element of the Bavarian Way was the provision of public capacities for applied research in conjunction with a publicly funded extension service. Other elements were the improvement of agricultural education systems and the promotion of cooperative formats like groups of producers.
The Bavarian State Research Center for Agriculture (LfL) was founded in 2003 in the course of a reorganization of 17 different units dedicated to research, education and other public tasks. The research station in Grub is today a hotspot of Bavarian animal breeding and includes the research station, an AI-organization, a veterinary service and research unit, a laboratory for routine genotyping and the head offices of several breeding organizations. Overall, there are more than 300 employees working for Bavarian animal breeders. The research in Grub covers all areas of animal production although only animal breeding will be described here.
The Institute of Animal Breeding of LfL
The Institute of Animal Breeding (ITZ) was founded as the department for animal genetics and IT in the 1970s under the lead of Gottfried Averdunk. Since 1990 the institute became responsible for genetic evaluations in Bavaria, and Averdunk was one of the initiators of Interbull in the 1980s. Animal breeding research at ITZ covers cattle, pigs, sheep/goats and horses, but no longer poultry. Grub used to host one of the last independent layer breeding programs in Europe, the so-called “Meisterhybriden”, until the selection unit was shut down in 1997 for “post-factual” reasons: the Bavarian government pushed the exit from conventional cages before the EU, and breed improvement without testing pedigree stock in cages seemed impossible. Lohmann acquired two of the brown layer lines in 1997 which became an ingredient of Lohmann “Tradition” [2].Apart from applied animal breeding research, the Institute of Animal Breeding of LfL (ITZ) runs a performance testing station for 4000 pigs/yr. and is responsible for conformation scoring in progeny of AIbulls (50.000 cows/yr.). It is also responsible for routine genetic evaluations in cattle, pigs, sheep and horses. Research cooperation exists with all German agricultural faculties and many others. A staff of ten senior researchers together with several PhD-students conducts applied research in quantitative genetics and the design of breeding programs.
Genetic evaluation in dualpurpose cattle
Cattle breeding has been a very open process for the last 70 years. Semen of bulls with high breeding values is traded worldwide and many AI-organizations in Europe and the US make a living from exporting semen to less developed countries. From the international exchange of semen and competition between AI-organizations in different countries, a need for comparable breeding values arose, which led to the foundation of “Interbull” and the development of the international genetic evaluation system for bulls known as MACE [3]. MACE supports the international trade of semen, but it does not offer the importing countries an opportunity to participate in international breeding programs, because it only provides “international” breeding values for AI-bulls, which may not adequately reflect the definition of traits in different countries. For the Fleckvieh breed we chose a different approach and developed an international genetic evaluation for bulls and cows giving comparable breeding values for all breeding animals and allowing for an across-country selection of sires as well as of (bull-)dams. Starting with a small national cooperation in 1995 that estimated breeding values for fat and protein yield, the genetic evaluation system for Fleckvieh (Simmental) and Braunvieh (Brown Swiss) was stepwise extended to more than 50 traits in breeding stock from Austria, Czech Republic, Italy, Hungary and Germany.The unique approach of the “Alpine Genetic Evaluation Team” (AGET) spreads the work load between three computing centers in Grub, Stuttgart and Vienna. Each team specializes on a specific group of traits and accumulates specific knowledge in this field. ITZ is focused on milk production and conformation traits, Stuttgart deals with the beef traits and Vienna specializes in reproduction, longevity and health traits. The scientists are coordinated by a technical committee and report to a transnational board of herd book organizations, AI-organizations and public servants. The basic idea behind the AGET approach is that it is relatively easy to extend a genetic evaluation to accommodate more data, while it is relatively complex to establish a genetic evaluation for a new trait.
Since 2009 a genomic evaluation system was developed and put into practice in 2011. AGET applies the same division of labor as for the conventional evaluations, thus allowing specific knowledge about the data in different trait groups to be used. The collaboration between the participating countries comprises not only the genomic evaluation, but logistics of DNA-samples, genotyping and genomic databases are shared in the same way. Conventional breeding values are published three times a year, while genomic breeding values are estimated monthly.
International trends in cattle breeding
The idea of sharing all the knowledge and breeding values across several countries and competing organizations may appear strange to pig and poultry breeders. However, this approach in cattle breeding has proven to be very successful in the past decades. The main reason for this was that performance testing of AI-bulls could only be organized using data from many (if not all) farms. Even non-herd book farms have contributed to performance testing and paid considerable shares of the recording system. Due to the necessity of progeny testing and the free distribution of semen, different breeding programs showed very similar rates of genetic progress. With the advent of genomic selection, the generation interval of a dairy cattle population began to depend heavily on the proportion of genomic young bulls used for AI. Varying proportions of young bulls lead to very different generation intervals and genetic progress becomes more heterogeneous across populations. In the long run the more progressive organizations will take the lead and will keep the best bull-dams of the breed for themselves, instead of sharing their genetic potential with all competitors. First tendencies can already be seen in the worldwide Holstein population [4]. Sexing Technologies, a relatively new player in the field of AI, uses promising new bulls exclusively for their own breeding program and later on some bulls are only available as sexed semen, not suitable for producing male calves. Other companies like Select Sires or Evolution distribute bulls to the general public only after a grace period of 4-6 months.This tendency is boosted by another trend in worldwide cattle breeding: For new traits, where phenotypes for historic breeding animals cannot be generated ex post, the genotyping of large cow samples is the method of choice. This requires large investments of the breeding organizations for genotyping and large efforts to supervise the participating farms. Breeding companies that have made these investments will not be willing to share the benefits with competitors. It is very likely that we will see a change of paradigm in the breeding of AI-bulls over the next few years. Cooperative breeding programs do not yet have developed a conclusive strategy for this new era.
Breeding of polled Fleckvieh
Most of the cows in present husbandry systems are kept in free stall systems, which require the cows to be hornless. However, the practice of dehorning causes pain for the animals and ethical concerns in the public. Polled breeds of cattle have been used since a long time, but polledness was usually not appreciated in multi-purpose breeds which were used for field work.Polledness is caused by a single locus which appears to be identical in all polled bos taurus breeds [5]. However, in most breeds used for dairy production polledness occurs only at very low frequencies, which results in a lack of polled AI-bulls and makes selection for polledness difficult.
Systematic breeding of polled Fleckvieh in Bavaria dates back until 1974 when the first polled cow was purchased for breeding purposes. Until 1985 three polled bulls and two more polled dams were purchased and from 1984 on the first homozygous bulls were produced via embryo transfer. Until 1990 the polled allele was only common in beef strains of the Fleckvieh breed. At the beginning of the 1990’s the first polled alleles were introduced into the dairy herd of the state research farm in Grub. This research farm acted as the nucleus for the introgression of the polled allele in the dual-purpose lines of the Fleckvieh breed. Since 2003 polled calves in all Bavarian herds under milk recording were systematically recorded and the polled phenotype was regularly examined by employees of ITZ. At the same time a systematic cooperation with Bavarian herd book organizations and AI-stations was established. Between 2000 and 2011 in total 193 polled bulls were raised at LfL, of which 37 entered AI-stations. The success of these bulls is also a nice example to demonstrate that a systematic application of quantitative genetic principles can lead to considerable progress even in species with long generation intervals.
Currently the dual-purpose population of Fleckvieh comprises 60 polled AI-sires and 15.000 polled cows. While the number of cows corresponds to 2% of the population, in 2016 already 18% of all inseminations were made with polled sires, 85% of them being young sires with only a genomic breeding value. The analysis of relationships of polled sires showed that the introgression has caused negligible amounts of additional inbreeding [6]. Fig. 1 shows the number of female polled calves registered by the Bavarian milk recording organization. Due to the incredible dynamics of the use of semen of polled bulls, it is very difficult to predict the share of polled cows in the future. However, from calves born in 2016 it is certain that the proportion will be close to 25% by 2021.
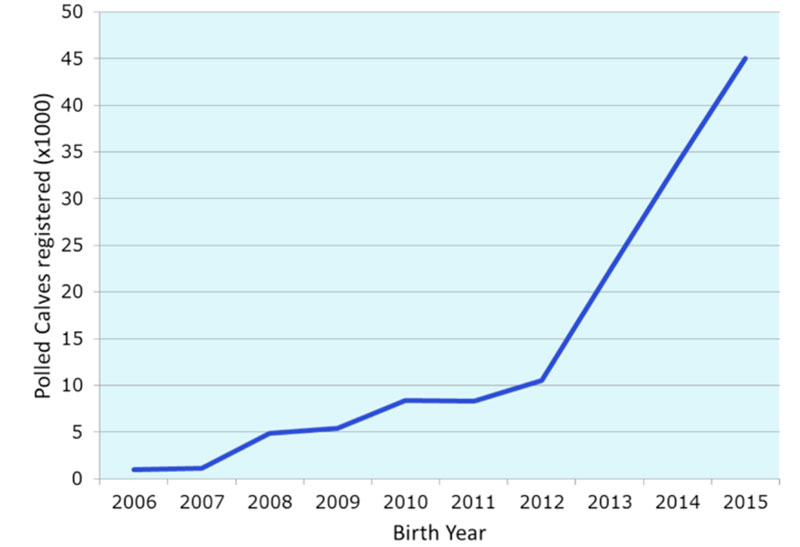
Figure 1 : Number of polled female calves born in herds under milk recording in Bavaria
Genomic Selection in Pigs
In Bavaria, herd book breeders are still supplying 79% of the genetic material used in piglet production, which is unique in Germany. Performance testing is very intense with approx. 300 newly recruited AI-boars per year providing 8.000 progeny tested on station. Until recently, genomic selection and routine genomic breeding value estimation has only been established in larger (international) pig breeding companies, while smaller breeding organizations are still working on the implementation, frequently struggling with small calibration sets. In Bavaria, a three-year project called InGeniS (“Integrierte genomische Forschung und Anwendung in der bayerischen Schweinezucht“ – integrated genomic research and application in Bavarian pig breeding) has been the kick-off for genomic selection in pigs. Within this project it was possible to genotype more or less all AI boars for the breeds Piétrain and German Landrace used as sires in Bavaria during the last decades with the Illumina PorcineSNP60 v2 BeadChip (around 60.000 markers). All these genotyped individuals build the basis for a routine genomic evaluation system which itself is the basis for applied genomic selection.Genomically enhanced breeding values (GEBV) can only be calculated if there is a sufficiently large set of individuals with both genotypic and phenotypic information that are used to calibrate the system, the so called reference set. These individuals are used to estimate the effects markers have on a specific trait. This will work out the better, the more individuals are available for the reference set, the better they represent the breeding population and the more own or progeny performance records are available. For the Piétrain breed in Bavaria there exist reference sets for around 2.500 individuals. This is one of the largest reference sets available for this breed and forms an excellent basis for the GEBV estimation.
Routine genomic evaluation in cattle normally involves a multi-step procedure with pedigree-based breeding value estimation, subsequent direct genomic breeding value estimation and finally a combination of both results [7]. A calibration set of many bulls with very reliable progeny-based breeding values is a good basis for such a procedure; however, such data is normally not available in pigs and thus small errors accumulate to a considerable amount across the different steps. Thus, a procedure called ‘single step’ is preferred for routine genomic evaluation [8]. This modelling is computationally more expensive, but provides the advantage of modelling all available information (phenotypic observations, pedigree data and genomic data for the genotyped individuals) in one single step, GEBV for all individuals directly.In pig breeding it is very common that a young selection candidate does not have own or progeny performance at the time of selection. Using conventional breeding value estimation the expected breeding value of such a candidate will be the mean of the parents’ breeding values. This parent average (with a reliability of around 20-30%, depending on breed) can only be used for selection between, but not within full-sib groups. The situation is similar to the one in layer breeding where early selection of males in the purebred lines is at a point in time where only phenotypes of full-sib sisters are available. In a genomic evaluation system, each selection candidate receives an individual GEBV from the moment its genotypic information is included into the single step model.
In Piétrain pigs in Bavaria reliability of the GEBV is approx. 50% compared to approx. 30% for a conventional parent average. With respect to the level of reliability, an early GEBV is comparable to the breeding value of a boar based on six crossbred progeny tested in the testing station. Apart from this increase in reliability the GEBV allows us to select within full-sib groups based on an individual breeding value for each member of the full-sib group. Figure 2 shows that clear differences between and within full-sib groups can be observed with GEBV. AI stations thus can use GEBV to decide which young boar to purchase and breeders can use GEBV to select the best females for their breeding stock.
Official genomic evaluation for pigs in Bavaria was introduced in May 2016 for the Piétrain breed and in December 2016 for German Landrace. Since then, GEBV are calculated for all individuals of the breeding populations on a weekly basis.
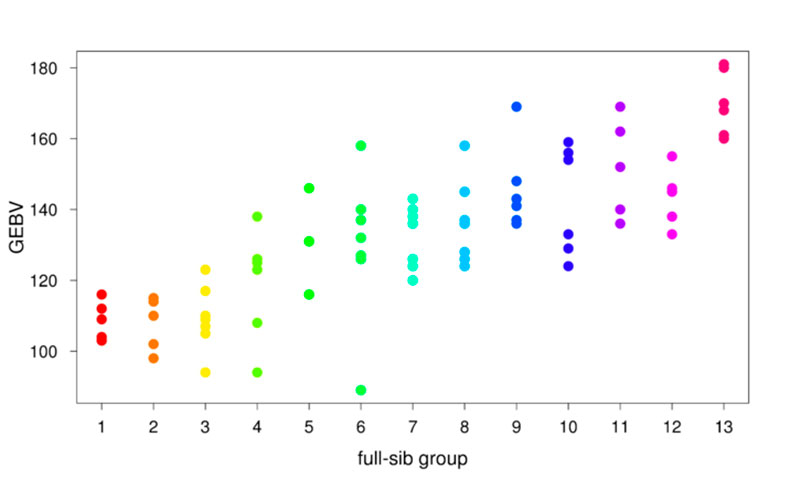
Fig. 2: Genomically enhanced breeding values (GEBV) for members of thirteen different full-sib groups from the Piétrain breed with at least six members. Each dot within in a full-sib group represents an individual GEBV of one of the members
Conclusions
The public research allows Bavarian farmers to compete in genetic progress and to apply state of the art methods although the majority of herd book and AI-organizations are way too small to run meaningful research units. Until now the Bavarian approach has proven to be resilient against purely market driven breeding programs. The availability of publically financed research allows also for the consideration of public goods in the breeding process. For example, meat quality traits in pigs have been considered since 1986, longevity, udder health and calving ease in cattle since 2002 and recently strong efforts have been made to prevent dehorning of cattle and tail-biting in pigs by means of animal breeding.

References
[1] Franke S (2012) Dr. Hans Eisenmann: Der bayerische Weg in der Agrarpolitik gestern und heute. http://www.hss.de/ download/120802_TB_Agrarpolitik.pdf[2] Preisinger R, Müller J, Flock DK (1999) Ökologische Eierproduktion aus züchterischer Sicht. Lohmann Information 2/99
[3] Schaeffer LR (1994) Multiple-Country Comparison of Dairy Sires. J Dairy Sci 77: 2671-2678
[4] Wesseldijk B (2015) Immer mehr Topbullen nicht mehr frei verfügbar. Holstein International 11/2015: 16-19
[5] Medugorac I, Seichter D, Graf A, Russ I, Blum H, Gopel KH, Rothammer S, Forster M, Krebs S (2012) Bovine polledness- an autosomal dominant trait with allelic heterogeneity. PLoS One 7, e39477
[6] Götz KU, Luntz B, Robeis J, Edel C, Emmerling R, Buitkamp J, Anzenberger H, Duda J (2015) Polled Fleckvieh (Simmental) cattle – Current state of the breeding program. Livest Sci 179: 80-85, http://dx.doi.org/10.1016/j.livsci.2015.05.019.
[7] VanRaden P (2008) Efficient Methods to Compute Genomic Predictions. J Dairy Sci 91:4414-4423
[8] Aguilar I, Misztal I, Johnson DL, Legarra A, Tsuruta S, Lawlor TJ (2010) Hot topic: a unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J Dairy Sci 93:743–752










