Summary
Well-balanced microbiota and subsequently a healthy gut have a direct influence on the efficiency of
nutrient utilization and overall animal health. This effect is the result of a very complex and highly
interactive ecosystem within the intestine formed by the intestinal microorganisms, host cells and diet
components. The experimental inaccessibility of many of these interactions, largely unknown microbiota
composition as well as a multiplicity of possible stable and “healthy” microbial community compositions
and processes complicate and hamper the definition of gut health. Therefore, a conclusive and
generally-accepted definition of these terms has not been found to date.
Nevertheless, several definitions for gut health and microbiota stability have been attempted in the past as well as measurable indicators for these conditions proposed. The indicators aim at characterizing the intestinal wall (e.g. villous height, crypt depth) and its functionality (e. g. mucus composition, permeability), the immune system (e.g., CD4 & CD8 cell numbers and their ratio) and the composition (e.g. number of beneficial and harmful organisms, genetic fingerprinting, diversity indices) and function (e.g. metabolite concentration) of the intestinal microbiota.
However, these indicators can not be regarded as principally applicable or by themselves sufficient to define gut health or microbiota stability. To influence microbiota stability and gut health systematically with feed additives requires the characterization of microbiota composition and representative indicators of gut health more specifically
Nevertheless, several definitions for gut health and microbiota stability have been attempted in the past as well as measurable indicators for these conditions proposed. The indicators aim at characterizing the intestinal wall (e.g. villous height, crypt depth) and its functionality (e. g. mucus composition, permeability), the immune system (e.g., CD4 & CD8 cell numbers and their ratio) and the composition (e.g. number of beneficial and harmful organisms, genetic fingerprinting, diversity indices) and function (e.g. metabolite concentration) of the intestinal microbiota.
However, these indicators can not be regarded as principally applicable or by themselves sufficient to define gut health or microbiota stability. To influence microbiota stability and gut health systematically with feed additives requires the characterization of microbiota composition and representative indicators of gut health more specifically
Introduction
The concepts of stability of the intestinal microbial composition and a therewith inferred so-called gut health seem to be increasingly popular. The search terms “healthy gut”, “gut health” and their synonyms, respectively, are cited more than 6000 times in the databases pubmed and Google scholar, which represents an exponential increase since their first mentioning in 18291 and 19132, respectively. Hence, within the scope of this article one might assume that the wide use of the respective terms implies a simple and well-established definition. Surprisingly, up to now a conclusive and generallyaccepted definition of these terms has not been found. One might even get the impression that not one serious attempt has been made. Furthermore, it is interesting that the mere acknowledgement of this unsatisfactory situation and a thorough discussion of the problems arising due to this fact are more or less absent in the scientific literature. Therefore, the frequent use of the concept of a stable microbiotadependent gut health is often quite uncritical and inaccurate and sometimes even careless.This article is an attempt at an approach to these concepts using three central questions:
(1) Why is gut health and microbiota stability important at all;
(2) How can these concepts be defined; and
(3) How can they be measured?
Importance of gut health and microbiota stability
First of all, both concepts have found their way into the European regulatory framework (EC No. 1831/2003), which regulates the use of feed additives. Even more importantly, well-balanced microbiota and subsequently a healthy gut have a direct influence on the efficiency of nutrient utilisation and overall animal health.The regulation EC 1831/2003 describes the functional group of “gut flora stabilisers” as microorganisms or substances, which have a “positive effect on the gut flora” of animals and thereby “favourably affecting animal production, performance or welfare”. Within this functional group an actual stabilisation is literally not necessarily the aim of the feed additive, but any effect on the microbiota deemed positive will suffice. Obviously, this is a rather vague definition and naturally the nature of the effects is the subject of controversial discussions. Nevertheless, for the topic of this article it should be noted that the regulation clearly states that microorganisms have an effect on welfare including the health of the animal.
This effect is the result of a very complex and highly interactive ecosystem within the intestine, which is formed between the consortium of all microorganisms present, host cells mainly of epithelial and immunological nature and diets of various compositions. The nature of the interaction between all components of this system is very difficult to describe both theoretically and experimentally. Microbiological factors, such as metabolite production, epithelial adhesion, pathogenicity factors, and nutrient utilization are an integral and indispensable part of this environment-host-interface which is formed on the other hand by the innate and acquired immune system, the mucous, epithelial surface factors, and the epithelial turnover rate. The efficiency of nutrient utilization in the small intestine is not only an economic issue in animal production but also important for the state of health of the individual animal. According to Anderson (2002)3 the effects of the intestinal colonization can be characterized as mainly competitive to host interest in the small intestine and mainly cooperative in the large intestine. The main competitive factors are concurrence for dietary energy and nitrogen sources as well as the resulting production of harmful microbial metabolites. The main cooperative factors are the provision of energy by microbial utilization of nutrients which are indigestible by host enzymes as well as the competitive exclusion of pathogens. The extent of the competitive and cooperative effects depends on the composition of the intestinal microbial community, which is therefore a rewarding and frequently aimed at target for intervention strategies.
The comparison of conventional and germfree animals demonstrates how important the intestinal microbiota are for the physiological development of the host. Very extensive discussions of the differences are found, for example, in articles of Berg (1996)4, Collinder (2001)5 and Tannock (2001)6 respectively. Amongst other things, microorganisms are responsible for morphological changes (e.g. larger intestinal surface, higher cell turnover, smaller caeca, faster peristalsis, higher mucin content, larger spleen and lymph nodes), immunological changes (more systemic γ-globulins, more intraepithelial lymphocytes, more IgA producing lymphocytes, faster immunological reaction to antigens, inducible oral tolerance) and metabolic changes (e.g. higher amount of SCFA, bile acid and bilirubin degradation, cholesterol utilization, β-glucuronidase and urease activity). It is obvious that all these changes together play an important role for the health of the host.
Although it is clear that microorganisms are responsible for all these changes, most species remain to be identified. Furthermore, besides effects caused by the collectivity of all microorganisms and in addition to the fact that microorganisms are present at all, species and strain-specific effects as well as the composition of the microbial community are important for the nature of the effects.
In the GI tract a delicate and tightly-regulated homeostasis between intestinal microorganisms and the immune system exist.7 A model put forward by Noverr et al. (2004) might serve as an example for a specific beneficial effect of the intestinal microbiota on health.8 The proposed model describes how the gastrointestinal (GI) microbiota might even regulate pulmonary immune responses in the airways to inhaled antigens that are non-infectious (pollens) or of extremely low infectivity (mould spores). The key roles in this model of oral tolerance have regulatory T-cell networks. Under non-inflammatory conditions, swallowed antigens are acquired by immature dendritic cells in the GI tract, which then promote the development of a regulatory T-cell network and prevent the development of over-exuberant Th2 responses.
On the other hand, certain long-term alterations in the bacterial and fungal GI microbiota induced through antibiotic use, dietary changes or illness as well as the total absence of GI microbiota as in germfree animals would alter the developmental environment for regulatory T cell responses. Amongst other things, antibiotics lead to overgrowth of the yeast Candida albicans which can secrete potent prostaglandin-like immune response modulators from the microbiota. Under conditions of particular microbiota alteration mature dendritic cells are primed upon encounter of non-infectious/low infectious microorganisms and an inflammatory Th2 response initiated, which leads eventually to allergy. Both IL-4 and IL-13 are involved in perpetuating Th2 responses.
In addition to the development of oral tolerance, “healthy” gut microbiota promote e.g. the IFN-γ induced gene expression of indoleamine 2,3-dioxygenase (IDO), nitric oxide synthase 2 (NOS2), phagocyte oxidase (phox), natural-resistance-associated macrophage protein 1 (NRAMP1) and guanylate-binding proteins (GBPs), which are involved in the inhibition of bacterial, protozoal and viral replication.9 The production of IFN-γ itself is promoted by regulatory T-cell networks and inhibited by Th2 responses. Just to name one example, Rhee et al. (2005)10 report convincing experiments proving an IFN-γ dependent colonization resistance. They found an attenuated intestinal inflammatory response and a higher systemic bacterial burden of Salmonella typhimurium in pups (and adult IFN-γ knockout mice) compared with adults, which were caused by an age-dependent shortcoming in the intestinal expression of a number of IFN-γ-regulated genes involved in antimicrobial defence. The developmental up-regulation of the IFN-γ-regulated genes was dependent on both IFN-γ and a normal commensal microflora.
However, it has to be considered – as stated by Cebra (1999)11 in a review of the influence of gut commensals on colonization resistance – that the underlying mechanisms are probably (and in my opinion very definitely) complex and various. For example, experiments of Lawley et al. (2008)12 show that antibiotic treatment causes a higher susceptibility to the transmission of Salmonella enterica Serovar Typhimurium through disturbance of the indigenous intestinal microbiota. Very similar observations were also reported by Sekirov et al. (2008)13. In their experiments antibiotics had a dosedependent effect on the intestinal microbiota composition although they did not significantly alter the total numbers of intestinal bacteria. Greater pre-infection perturbations in the microbiota resulted in increased susceptibility in mice to Salmonella serovar Typhimurium intestinal colonization, greater post-infection alterations in the microbiota, and more severe intestinal pathology.
Another aspect of the complex intestinal ecosystem is reported by Barman et al. (2008)14. They suggest that Salmonella infections in mice involved local mucosal host responses which take place prior to a disruption of the intestinal ecosystem. From alterations in microbiota composition which preceded the onset of diarrhoea in their experiments, they concluded that pathogen-commensal interactions and/or host responses unrelated to diarrhoea are involved.
Definition of gut health and microbiota stability
As suggested above, gut health and microbiota stability are very difficult to define due to the complexity of the underlining concepts. The closest approximation to a definition of gut health is the orientation at the WHO (1946)15 definition of general health itself: “Health is a state of complete physical, mental and social well-being and not merely the absence of disease or infirmity“. This definition is also the best indication for the difficulties encountered when defining “gut health” since although not very precise it has not been amended since 1948. At least, deduced from the health definition, “gut health” should include not only the complete histological intactness of the intestinal mucosa but also the overall functional “well-being” of the intestinal ecosystem, i.e. the optimal performance of its nutritional and immunological duties for the overall well-being of the host. When reviewing the literature this additional meaning of health explains the historical differences between the terms “gut health” and “healthy gut”, with the latter term appearing first in the literature of the early 20th century and relating initially only to the morphological intactness of the intestine while the former covers in its most frequent use all parts of the aspired definition. Nevertheless, it must be noted that both terms have been used so far in an inconsistent, versatile, ambiguous or at least unspecific way. Therefore, the definition of “gut health” remains unclear and is mainly determined by its ongoing use in the literature. Examples for this habitual use will be described in a later paragraph.In this context, the term “healthy” is often also used to describe the state of the intestinal microbiota. For example, eubiosis was defined as “stable and healthy microflora in the digestive tract”.16 The “stability” mentioned is expected to preserve a composition of certain microbial populations, which is often believed to be positive for “gut health”. Although “stability” and “dynamics” are antonyms in their colloquial use it must be noted that in ecological science “stability” is by no means a rigid, constant or everlasting condition. Botton et al. (2006)17 define stability as the ability of a system “to return to an equilibrium state after a temporary disturbance”. Consistently, eubiosis has been defined as “a state of a dynamic equilibrium of the microflora in stomach and intestinal tract”.18 The term dynamic equilibrium/ecological balance/biocoenotic balance describes the dynamic interrelationship of a biocoenosis (in open systems), which despite fluctuations in population frequency of single species or other disruptive factors (i.e. abiotic environmental factors) maintain the stability of the overall system as long as no principle milieu alteration occurs.19 If a stress situation cannot be compensated, this results in the succession into another ecosystem which might be as stable as the former one. Therefore, stability as such implies no assessment of the quality of the system, i.e. „healthy“ or „dysbiotic“. The biocoenosis is not in equilibrium during the succession so that the term stability is not expedient during this period.
The ability to react to constantly changing environmental conditions refers to both the composition (structural component) and the function (process component) of the system. According to Botton et al. (2006)17 and references therein, the stability of each of these components is usually not related and greater structural stability might even lead to less stability of function due to lower functional redundancy.
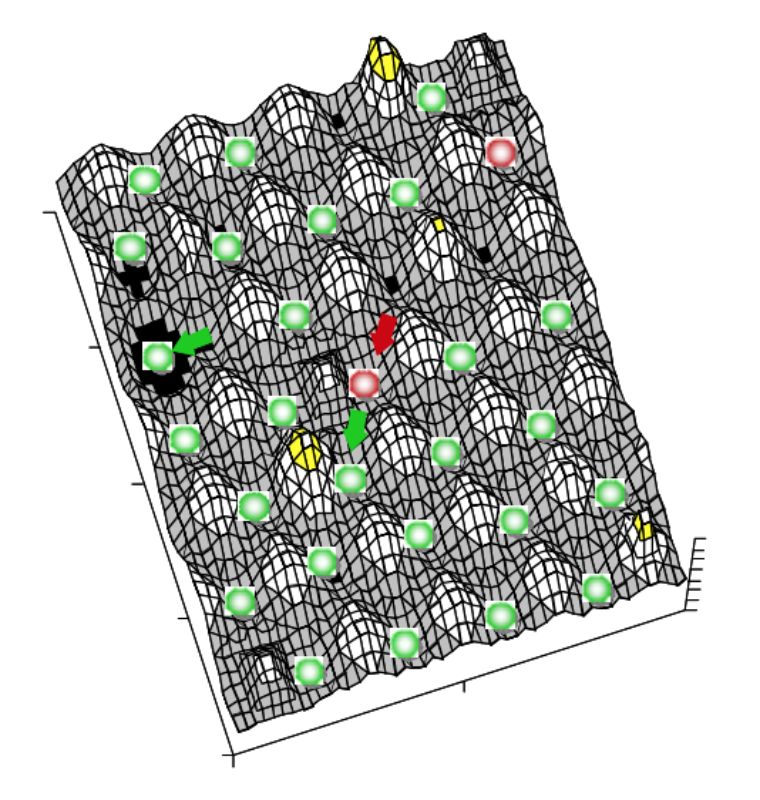
System stability can be depicted using a ball and cup heuristic, where valleys represent stability domains and balls the system.17 Resilience of the system in this model is described as slopes of the landscape and width of the valleys. Disturbances would push the system up the slope and ecological change may modify the shape of the landscape (i.e. change resilience height or modulate equilibrium points). For details see the excellent review of Botton et al. (2006)17. A three dimensional representation of this landscape model is given in Figure 1. In the intestine several different stability domains exist, each possibly with different domain slopes regarding their valleys and slope shape, i.e. resilience characteristics. The form of the stability domain and the character of the microbial
system, and consequently the immense number of possible stable systems will depend on the age of the host, the kind of host species and genotype, secretory products of the host, peristalsis, the type of diet (structure, viscosity, components), the environmental condition (e.g. management and housing), appliance of therapeutics, confounding illnesses and the kind of intestinal segment. Many of these stable systems might be characterized as “healthy” microbiota and just a few as “dysbiotic” conditions. Nevertheless, this multiplicity of possible “healthy” communities and processes complicates and hampers the establishment of a definition of gut health.
Figure 1: Three dimensional representation of
a possible section of a landscape
model of system stability within the
gut. Green balls depict “healthy”
microbiota and red balls “dysbiosis”.
Valley width and slope steepness
characterize resilience of the system.
The landscape shape (system
characteristics) varies in the intestine
depending on several factors (see
body of the text).
In addition, there are not only several possible „healthy” microbiota compositions, but also the analysis and characterization of the composition of the intestinal microbiota is methodological difficult if not in its entirety completely impossible.
A very simplified and surely too naïve idea of microbiota characterization is its classification into taxa (mostly on the genus level) with “harmful effects” on one side and those with “health promoting functions” on the other side.20 In these models Clostridium spp. are placed univocally in the former category while Lactobacillus spp. are placed in the latter. These models are problematic due to their generalization and simplification. Tannock (2003)21 reviews several reports on exceptions of this simplified classification: Lactobacilli (inter alia L. rhamnosus, L. reuteri) as well as other Probiotic organism containing genera can cause bacteraemia, rarely with localisation on heart valves or liver abscess. Nevertheless, the bacteraemia is most often only caused together with underlying conditions such as cancer, diabetes mellitus, or recent surgery and is often polymicrobic.22-28 Quite similarly, Saccaromyces boulardii strains might cause fungemia.29-31 In addition, potentially harmful antibiotic resistance can be found in several Lactobacillus and Bifidobacterium strains.32-33 It should also has to be noted that Clostridiacea and Enterobacteriacea can not be generally classified as harmful. Butyrate production in the large intestine by Clostridium spp. is an important ecological function34-38 and Escherichia coli and Salmonella strains have probiotic functions.39-44
A further model divides the microbiota into “main flora”, which consist of the most common gram positive and/or gram negative bacteria (i. e. Lactobacillus spp., Bifidobacterium spp., Bacteroides spp., Eubacterium spp.), the “secondary flora” (i. e. Enterococcus spp., E. coli) and the “residual flora” (potentially pathogenic Proteus spp., Staphylococcus spp., Pseudomonas spp.). The state is referred to as „eubiosis” if the ratio of main:secondary:residual flora is 90:1:0.01.45 This classification is also problematic in at least two aspects. By concentration on group frequencies it ignores that very low counts of pathogens might cause illnesses without changing the proposed ratios of main, secondary and residual flora. In addition, shifts in species composition within these groups will not be noticed although sufficient to cause either beneficial or harmful effects.
To get a more realistic picture of the complexity of the intestinal microbiota, the following facts should be recognized why it is so difficult to characterize the intestinal microbiota in detail and why simple classification approaches are insufficient:
• The intestine contains about 1014 bacteria, which to use a common comparison is more than the number of humans who have ever lived.
• The intestine harbours at least 10 Phyla in at least 34 families and 51 genera. Estimation of species richness range from about 1.000 up to as much as 16.000 species, which might have to be subdivided into more than 30.000 subspecies.46-48
• An estimated 20–90% of phylotypes (depending on habitat) are thought to have eluded cultivation so far and it is estimated that from at least 20% of phylotypes not even DNA sequences are known.46, 48-50
• high variability and heterogeneity:
a) Inter-individual: host specificity, genetic relatedness, environmental variation (region, diet, age etc.), health status.46, 49, 51-53
b) Intra-individual: site-specific distributions lead to differences between body regions, proximal:distal intestinal sites and luminal:mucosal sites, temporal variation.49-50 For example, in chickens about 70-90% of sequences from the small intestine are related to those of Lactobacillus spp.. In contrast, Clostridiaceae-related sequences (60-80%) are the most abundant group detected in the cecum.54-57
• Taxonomic diversity and functional diversity are not congruent with each other.
Proposed indicator for gut health and microbiota stability
From the previous paragraphs it has to be concluded that no tangible, generally-accepted and applicable definition of both, gut health and microbiota stability exists so far. Nevertheless, several feed additives claim effects on both variables and support this by studies investigating what might be called “soft” indicators (e.g., overall health and performance, absence of pathogens, abundance of GRAS organisms, high colonic butyrate concentration, high diversity, quotients of selected microbial groups, immunological variables, etc.). All of these indicators have in common that they have been correlated somehow to both gut health and microbiota stability, but evidence of a clear dependency is missing. In addition, most studies cover only one or very few of these indicators so that these indicators can not be correlated conclusively among each other and a true countercheck of their effects remains missing. The use of these indicators arises more or less from tradition and a subjective impression of the gut ecosystem rather than being based on established facts. Nevertheless, recently available methods of measuring “normal” function of the gut and immune system as well as a “healthy gut flora” have been reviewed and definitions for these conditions attempted.58 Unfortunately, this review confines itself to the human ecosystem and remains incomplete. Some commonly used indicators shall be presented here exemplary.Some investigators look into the concentration of so called beneficial and harmful genera, respectively. For example, Pierce et al. (2006, 2007)59-60 conclude in studies with weaned piglets that “the inclusion of high dietary concentrations of lactose resulted in improved intestinal health through (..…) increases in lactobacilli (..…)”.The problems associated with this indicator have already been discussed.
Other reports concentrate on the investigation of the composition of the total or dominant microbial community using techniques such as G + C profiling, Fluorescence- in situ-hybridization and genetic fingerprinting. Using G + C profiling Apajalahti et al. (2007)61 compared two broiler farms, one inconspicuous control farm and one farm with high Campylobacter prevalence. In caecum samples of the affected farm they observed a higher frequency of Lactobacillus spp. and a lower frequency of species of the Clostridium cluster IV. In addition, the Campylobacter burdened farm showed higher interindividual variations. Since these observations were in accordance with earlier investigations of this work group, they judge similar deviations from an average “normal” G + C profile as destruction of gut health.
Vaahtovuo et al. (2007)62 found a positive correlation between a Microbial Balance index (MBI) and performance (but not health) of weaner and grower pigs. In this index, which divides numbers of Bifidobacterium spp. and Faecalibacterium prausnitzii by numbers of the Bacteroides–Porphyromonas– Prevotella-group and the enteric group, we meet again the simplified classification of microbiota in harmful and beneficial taxonomic groups of higher order.
Botton et al. (2006)17 recently reviewed the literature on the correlation between microbial diversity and stability of the ecosystem function. The first references to stability due to higher biodiversity by Darwin (1859)63 and Mac Arthur (1955)64 were subsequently experimentally confirmed by others. This stability-diversity relationship concept is used, for example, by Scanlan et al. (2006)65, who detected higher diversity and temporal stability in fecal samples of the control group compared to Crohn’s patients based on Denaturing Gradient Gel Electrophoresis (DGGE). However, often a straightforward diversity-stability relationship does not exist. Ecosystem function and stability are more directly related to functional diversity than to taxonomic diversity.17 Higher species diversity might indirectly support functional stability of an ecosystem (redundancy and insurance hypotheses) due to functional overlap of different species. Further irregularities in the stability-diversity relationship, which go beyond the scope of this article, are discussed by Botton et al. (2006).
Therefore, other applied indicators approach the intestinal microbiota not solely on the basis of their taxonomic composition but also through their functional characteristics such as metabolite production. Pierce et al. (2006, 2007)59-60 believe that an increase in lactic acid production and short chain fatty acid concentration accompanied by a reduction of intestinal pH “may delay the multiplication of pathogenic bacteria thereby improving gastrointestinal health”.
Kuzmuk et al. (2005)66 detected in dogs receiving an animal-product based diet higher intestinal butyrate concentrations but also higher ammonia concentrations than in dogs receiving a plant-product based diet. Summarizing they concluded regarding the connection to gut health that butyrate was “shown to play a central role in maintaining the intestinal mucosal barrier. In contrast, (…..) ammonia is a putrefactive compound that induces faster turnover of epithelial cells and is toxic to colonocytes.” In the same study villous height and crypt depth are described as “direct representations of the intestinal environment and may be used as indicators of intestinal health”.66 Ammonia has been negatively correlated to reduced villous height by disturbing the development of the intestinal mucosa”.59
In accordance with the indicators presented so far, Jeurissen et al. (2002)67 conclude that the health status of the intestine is determined by both characteristics of the microorganisms and characteristics of the intestinal wall. Their review describes indicators for immunity (e.g., CD4 & CD8 cell numbers and their ratio, immune cell function), integrity (villous height, crypt depth, expression of Ecadherin or trefoil factor) and functionality (permeability, mucus composition and secretion level) of the intestinal wall as characteristics of intestinal health. These authors believe that using a combination of these indicators “an accurate and detailed insight into the intestinal health of poultry can be obtained”.
Further indicators used to characterize gut health are gene expression analysis. For example, van Hemert et al. (2003)68 compared healthy chickens with those affected by the malabsorption syndrome (MAS). They found up-regulated genes in MAS affected intestine (e.g. lysozyme G and two interferon induced proteins) as well as down-regulated genes (apolipoprotein B, calbindin, and cytochrome). They conclude that “because Malabsorption syndrome (MAS) mainly affects the intestine, it can be used as a model to study intestinal health and intestinal disturbances in young broilers”.
Another indicator proposed is the intestinal fatty acid binding protein (I-FABP).69-71 Niewold et al. (2004) state that “in pigs circulating I-FABP is a useful marker for (mild) intestinal injury, and could possibly be used to monitor (intestinal) health in clinical practice”.71
The use of other indicators of gastrointestinal health (e. g., Cyclooxygenase-2, Citrulline) – sometimes only for the characterization of very special conditions- is discussed in the literature but their complete description is beyond the scope of this article.72-73 The most frequently-used indicators are cited here as an example for their diverse nature and to illustrate that from a single indicator no conclusive proof of the gut health level can be expected.
Conclusion
It has been demonstrated that “healthy” intestinal microbiota are essential to ensure the mutually beneficial interaction between microbiota and host as well as his diet, which affect a multitude of metabolic, morphological and immunological processes. Nevertheless, the composition of such “healthy” microbiota remains an open question. This is problematic since a specific and purposeful manipulation or even optimization of the intestinal microbiocoenosis is impossible without detailed knowledge of the composition and function of the microbial communities within the intestine as well as their conclusively established correlation to animal health and performance. Therefore, further efforts should be undertaken to characterize the composition as well as the function of the intestinal microbiota in conjunction with accompanying epidemiological investigations to eventually form a precise, generally accepted and coherent definition of gut health. Thereby even the opportunity will arise to identify indicators which have a prognostic significance with respect to the future development of gut health within one animal or flock.1. Abercrombie, J (1829). Amer J Med Sci 7: 123-165.
2. Briscoe, JF (1913). Proc R Soc Med 6(GenRep): 364-367.
3. Anderson, DB (2002). Proc.Elanco Global Enteritis Symposium, B1-B18.
4. Berg (1996). Trends in Microbiology 4(11): 430-435
5. Collinder, E (2001). Thesis, Karolinska University Press, Stockholm, Sweden
6. Tannock, GW (2001). Am J Clin Nutr 73 (suppl): 410S-414S
7. Kelly, D, S Conway and R Aminov (2005). Trends in Immunol 26 (6): 326-333
8. Noverr, MC and GB Huffnagle (2004). Trends in Microbiology 12(12): 562-568
9. Taylor, GA, CG Feng and A Sher (2004). Nature Rev Immunol 4: 100-109
10. Rhee, SJ, WA Walker and BJ Cherayil (2005). J Immunol 175(2): 1127-1136
11. Cebra, JJ (1999). Am J Clin Nutr 69 (suppl):1046S–1051S
12. Lawley, TD et al. (2008). Infect Immun 76(1): 403-416
13. Sekirov, I. et al. (2008). Infect Immun 76(10): 4726-4736
14. Barman, M. et al. (2008). Infect Immun 76 (3): 907-915
15. WHO (1946). Official Records of the World Health Organization, no. 2, p. 100
16. Wenk (2003). Alltech Petfood Seminar, 27.03.2003, Coventry, UK
17. Botton, S. et al. (2006). Crit Rev Microbiol 32(2): 101-112
18. Busch, A. et al. (1999). AWT, Agrimedia: 1-42
19. Schaefer, M and W Tischler (1983). Gustav Fischer Verlag, Stuttgart
20. Gibson, GR and MB Roberfroid (1995). J Nutr 125: 1401-1412
21. Tannock, GW (2003). Curr Issues Intest Microbiol 4: 33-42
22. Husni, R.N. et al. (1997). Clin Infect Dis 25: 1048-1055.
23. Rautio, M. et al. (1999). Clin Infect Dis 28: 1160-1162.
24. Woo, P.C.Y. et al. (2002). J Clin Microbiol 40: 265-267.
25. Farina, C. et al. (2001). J Clin Gastroenterol 33: 251-252.
26. Wagner, R.D. et al. (1997). Infect Immun 65: 3345-3351.
27. Harty, D.W. et al. (1994). Int J Food Microbiol 24 (1-2): 179-189
28. Salminen, M.K. et al. (2002). Clin Infect Dis 35: 1155-1160
29. Bassetti, S. et al. (1998). Am J Med 105 (1): 71-72
30. Cesaro, S. et al. (2000). Supp. Care Cancer 8 (6): 504-505
31. Fredenucci, I. et al. (1998). Clin Infect Dis 27: 222-223
32. Charteris, W.P. et al. (1998a). Int Dairy Technol 51 (4): 123-136
33. Charteris, W.P. et al. (1998b). J Appl Microbiol 84: 759-768
34. Varel, V.H. et al. (1995). Appl Environm Microbiol 61 (3): 1116–1119
35. Duncan, S.H. et al. (2002). Appl Environm Microbiol 68 (10): 5186–5190
36. Duncan, S.H. et al. (2004). Appl Environm Microbiol 70 (10): 5810–5817
37. Schwiertz, A. et al. (2002). System Appl Microbiol 25: 46–51
38. Louis, P. et al. (2004). J Bacteriol 186 (7): 2099–2106
39. Neish, A.S. et al. (2000). Science 289: 1560-1563
40. Möllenbrink, M and E Bruckschen (1994). Med Klin (Munich) 89 (11): 587-593;
41. Wehkamp, J. et al. (2004). Infec Immun 72 (10): 5750-5758
42. Sturm, A. et al. (2005).Infect Immun 73 (3): 1452–1465
43. Duncker, S.C. et al. (2006). Vet Immunol Immunopathol 111: 239–250
44. Kleta, S. et al. (2006). J Appl Microbiol 101 (6): 1357-1366
45. Gedeck, B. et al. (1993). Arch. Tierernährung 44: 215-226
46. Bäckhed, F. et al. (2005). Science 307: 1915-1920
47. Curtis, TP and WT Sloan (2005). Science 309: 1331-1333
48. Frank, D.N.F. et al. (2007). PNAS 104 (34): 13780–13785
49. Dethlefsen, L. et al. (2007). Nature 449: 811-818
50. Eckburg, P.B. et al. (2005). Science 308: 1635-1638
51. Ley, R.E., D.A. Peterson and J.I. Gordon (2006). Cell 124: 837–848
52. Anderson, M.A. et al. (2006). Appl Environm Microbiol 72 (11): 6914–6922
53. Palmer, C. et al. (2007). PLoS Biology 5 (7): 1556-1573
54. Dumonceaux, T.J. et al. (2006). Appl Environm Microbiol 72 (4): 2815-2823
55. Walter, J. (2008). Appl Environm Microbiol 74 (16): 4985–4996
56. Lan, P.T.N. et al. (2002). Microbiol Immunol 46 (6): 371-382
57. Lu, J. et al. (2003). Appl Environm Microbiol 69 (11): 6816–6824
58. Cummings, J.H. et al. (2004). Eur J Nutr 43 (Suppl 2): II/118–II/173
59. Pierce, K.M. et al. (2006). Anim Sci 82: 311–318
60. Pierce, K.M. et al. (2007). Anim Feed Sci Technol 132: 267–282
61. Apajalahti, J and A Kettunen (2007). Avian Gut Function in Health and Disease, GC Perry (Ed.), 124-137
62. Vaahtovuo, J. et al. (2007). Livestock Sci 109: 174-178
63. Darwin (1859). The Origin of Species.
64. Mac Arthur, R. (1955). Ecology 36: 533-536
65. Scanlan, P.D. et al. (2006). J Clin Microbiol 44 (11): 3980–3988
66. Kuzmuk, K.N. et al. (2005). J Nutr 135: 1940–1945
67. Jeurissen, S.H.M. et al. (2002). Curr Issues Intest Microbiol 3: 1-14.
68. van Hemert, S. et al. (2003). Anim Biotechnol 14 (2): 133 – 143
69. Smolders, G. and M. Plomp (2007). Rapport 45, Animal Sci. Group Wageningen,
70. van Hemert, S. et al. (2004). Poultry Sci 83: 1675–1682
71. Niewold, T.A. et al. (2004). Res Vet Sci 77 (1): 89-91
72. Rhoads, J.M. et al. (2005). J Pediatr 146 (4): 542-547
73. Nurmi, J.T. et al. (2005). Nutr Cancer 51 (1): 83-92
Zusammenfassung
Das mikrobielle Gleichgewicht und die u. a. dadurch bedingte Darmgesundheit haben einen direkten Einfluss auf die Effizienz der Nährstoffverwertung und die Tiergesundheit. Diese Wirkung ist die Folge eines sehr komplexen und hoch interaktiven Darmökosystems aus Darmbakterien, Wirtszellen und Futterkomponenten. Die experimentelle Unzugänglichkeit vieler dieser Interaktionen, die zu einem großen Teil unbekannte Zusammensetzung der Mikrobiota sowie die Mannigfaltigkeit der möglichen stabilen „gesunden“ Zusammensetzungen mikrobieller Gemeinschaften und Prozesse kompliziert und behindert die Etablierung einer Definition der Darmgesundheit. Deswegen gibt es bislang keine schlüssige und allgemein anerkannte Definition der eingangs genannten Begriffe.
Nichtsdestotrotz gibt es viele Definitionsversuche für Darmgesundheit und Mikrobiota-Stabilität und einige messbare Indikatoren für diese Konzepte wurden vorgeschlagen. Diese Indikatoren charakterisieren den Zustand der Darmwand (z. B. Villushöhe und Kryptentiefe) und ihre Funktion (z. B. Mucuszusammensetzung, Permeabilität), das Immunsystem (z. B. CD4- & CD8-Anzahl und ihr Quotient) sowie die Zusammensetzung (z. B. Anzahl günstiger und gesundheitsschädlicher Bakterien, genetische Fingerabdrücke, Diversitätsindizes) und Funktion (z. B. Metabolitenkonzentrationen) der intestinalen Mikrobiota.
Allerdings können diese Indikatoren keineswegs als grundsätzlich anwendbar oder durch sich selbst hinreichend zur Definition von Darmgesundheit und Mikrobiota-Stabilität gelten. Um MikrobiotaStabiltität und Darmgesundheit durch Futterzusatzstoffe systematisch und gerichtet zu beeinflussen, wird es in Zukunft notwendig sein, die mikrobielle Zusammensetzung detaillierter zu charakterisieren und spezifische und wirklich repräsentative Indikatoren für die Darmgesundheit zu identifizieren.







