For the diagnosis of diseases in veterinary medicine the technique of PCR (polymerase chain reaction) could be used since
several years. In this method, individual genome sections of
the pathogen are propagated by repeating different temperature steps and thus finally made them detectable. In addition,
pathogens could be typed and differentiated from vaccine
strains. This can for example be done by a vaccine – specific
PCR or by subsequent sequencing of the PCR product.
The direct detection of the genetic material of the pathogen by means of PCR, in contrast to serological tests, has the advantage that diseases can be promptly detected after the initial infection. The PCR provides here a faster method than e.g. the cultivation of viral pathogens in eggs or cell cultures. However, the genetic material of the pathogen may possibly no longer be detected after a successful fight of the immune system against infection. Thus, the PCR may serve as a practical complement to serological tests that detect the antibodies produced by the host organism and help to overcome the gap in diagnostics between the time of infection and the production of antibodies.
Basically, PCR reactions could be differentiated in two techniques: During the first one, the “Endpoint” -based PCR, after the PCR reaction the products are separated in an agarose gel according to the size of the product and are stained. With the faster and more sensitive method the formation of the PCR products during the temperature cycles is observed in real time (Fig. 1). In recent years the veterinary laboratory has almost all the PCR detection methods converted to real-time PCR. This allows faster processing of the samples and more precise results by using this more sensitive method.
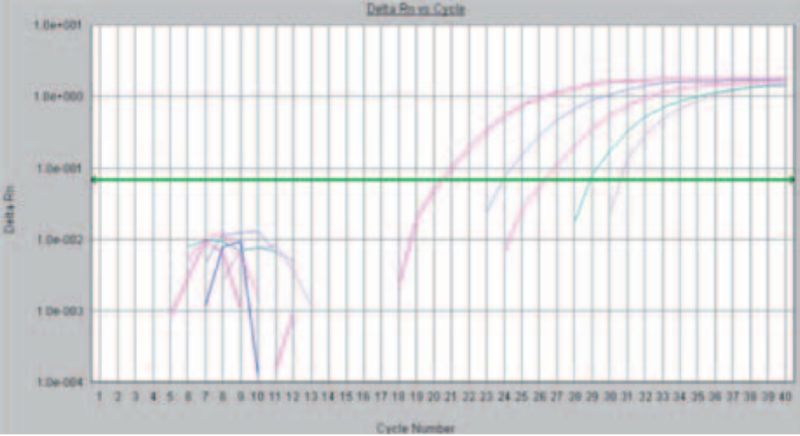
Figure 1: Detection of Avian Infectious Bronchitis via “real time” PCR. The rising curves indicate an increase in fluorescence and thus prove the presence of genetic material of the pathogen
Frequently requested examinations in the veterinary laboratory of LTZ are the detection of Avian Influenza (AIV), Avian Infectious Bronchitis (IBV), Mycoplasma (Mg / Ms) and Avian Metapneumovirus (AMPV, TRT). AIV can by typed by further PCRs in H5, H7 or H9. For serotyping of IBV recently “Real Time” detection systems for the serotypesQX, Italy O2, Massachusetts, D 274, 4/ 91 and D 1466 have been newly developed. By using a further PCR followed by sequencing positive Ms and Mg results can distinguish vaccine from field strains.
Further real-time PCR tests can for example be performed for the detection of Newcastle disease virus (NDV), Marek’s Disease and Egg Drop Syndrome virus (EDS). For ND virus a further PCR reaction followed by sequencing of the F1 cleavage site could determine pathogenicity and classification of the strains.
Overall, as established in the veterinary laboratory at present the detection of more than 25 pathogens is possible, with development of further detection systems. As matrices for the detection tracheal or cloacal swabs, FTA cards or organs can be used depending on the agent to be detected.
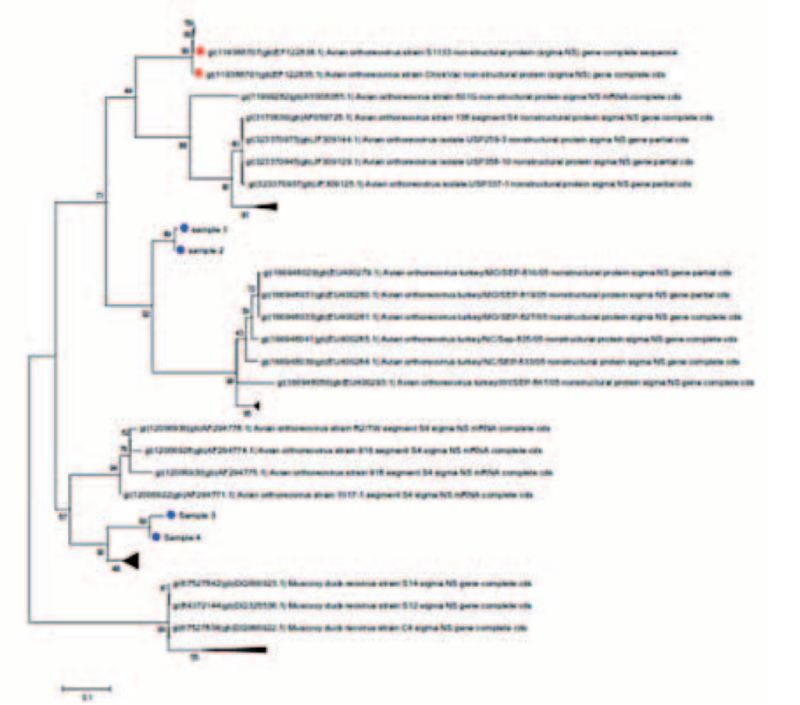
Figure 2: Phylogenetic trees of selected Orthoreoviruses; blue points are from field isolates, vaccine strains in red
In addition, typing can be carried out for Avian Orthoreovirus, Fowl Adenovirus (FAdV) and Infectious Bursal Disease Virus (IBDV). Through this, vaccine from field strains could be distinguished, the phylogenetic relationships of different virus isolates be determined (Fig. 2) and serotyping could be done. This method can serve as a decision aid in the selection of isolates for the production of autogenous vaccines. For typing of FAdV and Orthoreovirus the initial culture in liver cells is recommended in order firstly to achieve enrichment of the viral material as well as to be able, where appropriate, to use the isolate obtained for vaccine production later.
Dr. Martin Barz and
Dr. Matthias Voss,
Veterinary Laboratory
The direct detection of the genetic material of the pathogen by means of PCR, in contrast to serological tests, has the advantage that diseases can be promptly detected after the initial infection. The PCR provides here a faster method than e.g. the cultivation of viral pathogens in eggs or cell cultures. However, the genetic material of the pathogen may possibly no longer be detected after a successful fight of the immune system against infection. Thus, the PCR may serve as a practical complement to serological tests that detect the antibodies produced by the host organism and help to overcome the gap in diagnostics between the time of infection and the production of antibodies.
Basically, PCR reactions could be differentiated in two techniques: During the first one, the “Endpoint” -based PCR, after the PCR reaction the products are separated in an agarose gel according to the size of the product and are stained. With the faster and more sensitive method the formation of the PCR products during the temperature cycles is observed in real time (Fig. 1). In recent years the veterinary laboratory has almost all the PCR detection methods converted to real-time PCR. This allows faster processing of the samples and more precise results by using this more sensitive method.

Figure 1: Detection of Avian Infectious Bronchitis via “real time” PCR. The rising curves indicate an increase in fluorescence and thus prove the presence of genetic material of the pathogen
Frequently requested examinations in the veterinary laboratory of LTZ are the detection of Avian Influenza (AIV), Avian Infectious Bronchitis (IBV), Mycoplasma (Mg / Ms) and Avian Metapneumovirus (AMPV, TRT). AIV can by typed by further PCRs in H5, H7 or H9. For serotyping of IBV recently “Real Time” detection systems for the serotypesQX, Italy O2, Massachusetts, D 274, 4/ 91 and D 1466 have been newly developed. By using a further PCR followed by sequencing positive Ms and Mg results can distinguish vaccine from field strains.
Further real-time PCR tests can for example be performed for the detection of Newcastle disease virus (NDV), Marek’s Disease and Egg Drop Syndrome virus (EDS). For ND virus a further PCR reaction followed by sequencing of the F1 cleavage site could determine pathogenicity and classification of the strains.
Overall, as established in the veterinary laboratory at present the detection of more than 25 pathogens is possible, with development of further detection systems. As matrices for the detection tracheal or cloacal swabs, FTA cards or organs can be used depending on the agent to be detected.

Figure 2: Phylogenetic trees of selected Orthoreoviruses; blue points are from field isolates, vaccine strains in red
In addition, typing can be carried out for Avian Orthoreovirus, Fowl Adenovirus (FAdV) and Infectious Bursal Disease Virus (IBDV). Through this, vaccine from field strains could be distinguished, the phylogenetic relationships of different virus isolates be determined (Fig. 2) and serotyping could be done. This method can serve as a decision aid in the selection of isolates for the production of autogenous vaccines. For typing of FAdV and Orthoreovirus the initial culture in liver cells is recommended in order firstly to achieve enrichment of the viral material as well as to be able, where appropriate, to use the isolate obtained for vaccine production later.
Dr. Martin Barz and
Dr. Matthias Voss,
Veterinary Laboratory






